Data Finders
How can artificial intelligence and deep-learning platforms be used to combat malaria?
Despite all of effort and money that has gone into malaria research, increasing numbers of multi-drug resistant parasites are being reported, and the disease remains a significant global burden. Though several effective treatments have been developed, they fail to wipe out all of the parasites in hosts and are likely to encourage long-term resistance with selection pressure.
Optibrium – a provider of software and services for drug discovery; and Intellegens – a start-up focusing on AI, have collaborated on a predictive model that is able to identify antimalarial compounds. The new Alchemite-based platform was put to the test in the Open Source Malaria (OSM) global initiative, which invites scientists to share their research on malaria drugs through an online platform. The AI platform developed by the two companies uses novel mechanisms that could be critical to future malarial control and elimination. Here, we speak to Benedict Irwin, Senior Scientist at Optibrium, about the newly formed partnership and the role AI is playing in combating one of the world’s most deadly diseases.
How did you come to collaborate with Intellegens?
Both companies have strong links to the Theory of Condensed Matter (TCM) group in the Cavendish Laboratory at the University of Cambridge. Intellegens is a spin out company from TCM and our CEO, Matthew Segall, and I both did our PhDs in the group. It was during this time that we met Gareth Conduit, the CSO of Intellegens.
Intellegens’ initial research focused on material sciences; the team created Alchemite to help develop new super-alloys; in essence, the algorithm uses sparse and noisy data to make predictions. During discussions between Matthew and Gareth, it was quickly realized that the capabilities of the algorithm were ideally suited to applications in drug discovery; after all, our field produces a lot of sparse and noisy data, which Alchemite works well with! Furthermore, Optibrium’s extensive drug discovery domain knowledge had the potential to help Intellegens enter our sector through a collaborative partnership. The concept was further cemented with a grant from Innovate UK, which was used to fund a £1-million project called Deep ADMET (using deep-learning to predict the absorption, distribution, metabolism, excretion and toxicity of new drug candidates) in collaboration with the Medicines Discovery Catapult. We learned a lot from this project and other areas – and have since refined Alchemite.
How does Alchemite work?
Alchemite performs a process called “deep imputation,” which uses deep-learning algorithms to fill in the missing entries in a data set with predictions. The predictions can be based on structural descriptors, as well as any existing and/or measured datapoints themselves, even if most of the potential data have not yet been measured. Not only can the algorithm learn from structure-activity relationships, as used by conventional quantitative structure-activity relationship (QSAR) models, but also assay–assay correlations, which provide much more information to make more accurate predictions.
One of the other strengths of the method is that it provides an error-bar on every prediction, so we know the confidence in each one. It is impossible to get a perfect model of biological data, especially if there is a lot of noise in the original data, but with Alchemite we can focus on the accurate predictions hidden underneath the noise, regardless of the overall quality of the data. It is this feature that helped generate reliable predictions for active compounds for the OSM project.
How did you get involved with OSM?
Tom Whitehead, Head of Machine Learning at Intellegens, noticed the competition advertised on the OSM consortium’s online platform. OSM’s overall goal is to find new medicines for the treatment of malaria, and the competition was to develop computational models that could predict which molecules would most effectively tackle the malaria parasite. I’ve always been interested in the power of collaborative science and was keen to participate.
OSM developed a test set of a hidden batch of compounds, and had separate submissions for industry and academia. Our Alchemite model was selected and tested as one of the four winning models. Based on this preliminary success, we were invited to generate active compound suggestions which, in turn, OSM would synthesize. Excitingly, our first compound was found to be active.
Why is it so difficult to develop effective treatments and vaccines for malaria?
Plasmodium parasites have much more complex lifecycles than viruses or bacteria, including a dormant phase in liver cells and an active phase in the blood, during which clinical symptoms are observed. It is, therefore, critical to target a number of stages in the malaria lifecycle simultaneously during treatment to prevent entry into liver cells at the same time as treating the blood for the adult-stage parasites.
Medicines against malaria do exist, but resistance to some treatments has developed over time and the long-term use of medication is often unaffordable for those most affected by the disease. There is also a risk of severe side effects with some medicines.
In terms of a vaccine, there has been some great work but there are still concerns about efficacy. Previous malaria vaccines, such as SPf66, have failed to have the desired impact when used in patients.
The goal of OSM is to develop actives against a newer target protein in the cycle, PfATP4. Compounds targeting this protein are lethal to the parasite, but it is not yet clear how drugging this target results in the death of the parasite.
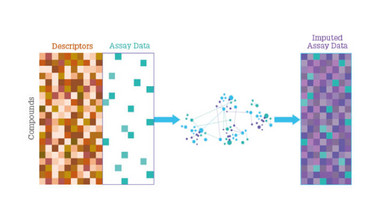
Figure 1: Schematic of a typical drug discovery dataset. Compounds are represented with descriptors (orange), which can all be calculated in advance. Only some of the experimental measurements have been performed (green), which gives a sparse, assay data matrix. Alchemite fills in the missing values using deep imputation.
Why is Alchemite suited to helping in the fight against malaria?
The target protein PfATP4 has no known structure, so investigation relies on phenotypic drug discovery, and machine learning models could prove to be useful tools in working out structure-activity relationships. Alchemite can identify correlations across different columns of sparse data in a data set and can learn from incomplete measurements for different strains, which are likely to correlate for some compounds and not for others. An example of this is depicted in Figure 1 – whereAlchemite was shown to be effective in identifying drug-resistant models of some of the most resilient strains of Plasmodium.
What’s the next stage for this project?
The results of our project will be submitted to the Beilstein Journal of Organic Chemistry, once the current batch of compounds have been fully documented. But in the longer term, the search will continue. Alchemite recently produced interesting data about the structure-activity relationship of compounds in a lead series; it suggested replacing a key group in some compounds with bulky tert-butyls. This prediction adds a new dimension of exploration for us.

Making great scientific magazines isn’t just about delivering knowledge and high quality content; it’s also about packaging these in the right words to ensure that someone is truly inspired by a topic. My passion is ensuring that our authors’ expertise is presented as a seamless and enjoyable reading experience, whether in print, in digital or on social media. I’ve spent fourteen years writing and editing features for scientific and manufacturing publications, and in making this content engaging and accessible without sacrificing its scientific integrity. There is nothing better than a magazine with great content that feels great to read.