QTOF: Quick Off the Mark for Oligonucleotide Analysis
Experts from Axolabs GmbH and Bruker Daltonics explain the benefits behind mass spectrometry for oligonucleotides in drug discovery
Timo Schierling, Julia Schneider, Stephan Seiffert, Christian Albers | | 4 min read | Technology

Oligonucleotides facilitate the rapid amplification of millions of DNA segments and have applications in the fields of biotechnology, diagnostics, forensics, and pharma – where they’re increasingly seen as one of the most promising therapeutic modalities.
Oligonucleotides are short synthetic strands of DNA or RNA that can be used to modulate gene expression. Antisense oligonucleotides (ASOs) specifically target the messenger RNA (mRNA) involved in the formation of disease-related proteins in the body; by binding to complementary oligonucleotides, they help with the detection, modification, restoration, or reduction of these proteins. As ASOs can replicate unknown gene expressions, neurodegenerative illnesses that have been linked to the formation of mistranslated proteins, such as Parkinson’s and Alzheimer’s, are particularly promising targets (1).
It is estimated that, as of 2020, over 50 ASOs were in clinical trials, with more on the horizon (2). However, the complexity of oligonucleotides makes them hard to purify – and any impurities or degradation can impact drug safety and efficacy. The key to gaining regulatory approval – and thus reaching patients – depends on the accurate analysis and characterization of the structural sequence of oligonucleotides. Specifically, the ability to assess the identity, purity, quality, strength, structure, and physicochemical properties of oligonucleotides is a prerequisite for transition from bench to bedside.
The sequence accuracy of RNA and oligonucleotides is critical to the characterization of impurities or degradation in the biopharmaceutical product approval process. From a clinical point of view, comprehensive characterization ensures the correct activity and expression. Importantly, this can help avoid toxicity or off-target effects for patients.
Why MS?
MS is considered by many to be the “gold standard” for oligonucleotide characterization, due to its enhanced sensitivity, robustness and mass accuracy (4). Various methods have been found to be effective in oligonucleotide analysis, though each has drawbacks. For example, electrospray ionization (ESI) allows the analysis of photosensitive and fragile oligonucleotides but can only handle 10-100 samples per day. Matrix-assisted laser desorption/ionization (MALDI) can process thousands of samples per day, but can only handle shorter sequences in the 20-30 nucleotides range (5).
Moreover, confirmation of mass is not always enough when it comes to checking for purity. Information at the nucleotide sequence level is often also needed for detecting impurities and any degradation pathways.
Tandem MS (MS/MS) provides the sensitivity, robustness and high dynamic range needed to detect these impurities. However, MS/MS spectra are complex; manual interpretation requires scientists with specialist, in-depth analytical skills, making full analyses both costly and lengthy.
A higher certainty
Quadruple Time of Flight (QTOF) MS instruments are widely used for characterizing highly modified oligonucleotide sequences as they are able to determine monoisotopic masses of intact oligonucleotides and their fragment ions with high mass accuracy. QTOF instruments, when combined with the right software, have positively impacted throughput. Moreover, software advances are resulting in powerful tools for the interpretation of RNA and oligonucleotide data, ensuring correct mass assignments even in samples covering a high dynamic range (see Figure 1).
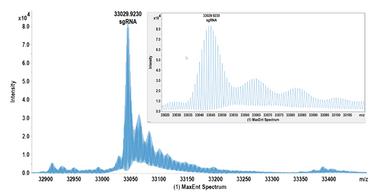
Figure 1. Deconvoluted spectrum for a sgRNA and associated low level impurities. Monoisotopic mass is annotated.
The SNAP algorithm together with high isotopic fidelity QTOF instrumentation enables the selection of the monoisotopic signal of one fragment, without isotopic ones, meaning spectra are matched with high certainty to sequence candidates. This leads to fewer mismatches. Current software offers algorithms and workflows to annotate MS/MS data from oligonucleotide sequences up to 100 nucleotides (1).
QTOF-MS has proven to be an effective technique for analyzing heterogeneities in oligonucleotides, such as small interfering RNA (siRNA), as well as in larger molecules. Indeed, high isotopic fidelity allows characterization of nucleic acid macromolecules, such as single-guide RNA (sgRNA).
Oligonucleotides of the future
With their ability to interact with protein targets and inhibit genes, oligonucleotides are increasingly in demand in fields such as diagnostics and pharma, especially in relation to diseases with unknown gene mutations.
To satisfy regulatory approval and smooth the path to market, accurate characterization is essential. In this regard, Axolabs has found that QTOF MS/MS allows faster, more accurate sequencing of oligonucleotides and RNAs, while recent software advances enable faster spectra interpretation with less manual interpretation required from laboratory staff.
What does this mean for drug development? We are living in what has been described as a “therapeutic revolution” with oligonucleotide and RNA therapeutics having “unlimited capacity to address unmet clinical needs.”(2) This could include the actualization of personalized medicine (3) and – particularly, pertinent in the aftermath of COVID – improvements in the efficacy of vaccines.
The right instrumentation and software will likely play a crucial role in bridging the MS/MS data gap and accelerating the process from research lab to the clinic – and as analysis and characterization become more accurate, oligonucleotides may be able to match or even surpass the success of small molecule and antibody drugs.
- Bruker Daltonics, Bruker Announces Makor Advances in CCS-Enabled 4D Proteomics at ASMS, 2021, https://www.businesswire.com/news/home/20211102005437/en/Bruker-Announces-Major-Advances-in-CCS-Enabled-4D-Proteomics-at-ASMS
- Damase T.R, The Limitless Future of RNA Therapeutics, Frontiers in Bioengineering and Biotechnology, 2021, https://www.frontiersin.org/articles/10.3389/fbioe.2021.628137/full
- Roberts T.C, Advances in oligonucleotide drug delivery, Nature Reviews Drug Discovery, 2020, https://www.nature.com/articles/s41573-020-0075-7
- Sutton, J.M, et al, Current State of Oligonucleotide Characterization Using Liquid Chromatography-Mass Spectrometry: Insight into Critical Issues, 2020, https://pubs.acs.org/doi/pdf/10.1021/jasms.0c00179#
- Anjali Alving, Bruker Daltonics, Simplifying Oligonucleotide Drug Development Workflows with Mass Spectrometry, 2019, https://www.bruker.com/en/news-and-events/webinars/2019/simplifying-oligonucleotide-drug-development-workflows-with-mass-spectrometry.html
Scientist (Analytics R&D)
Principal Research Associate
Team Leader (Method Development Analytics), Principal Scientist, Axolabs GmbH
Business Development Manager, Pharmaceuticals Market, Bruker Daltonics