Making Sense of Change
Why serological testing will play an essential role in COVID-19 surveillance and vaccine monitoring
Andy Lane | | Longer Read

Andy Lane is the Commercial Director at The Native Antigen Company.
As the number of cases of COVID-19 began to climb in late 2019, labs began collecting and sequencing patient isolates. Within weeks, the first draft sequence of the SARS-CoV-2 genome was published, opening the gates for a wave of early research (1). At the time, one of the major questions surrounding SARS-CoV-2 was its natural mutation rate. It has long been understood that viral genomes naturally mutate over time. However, the pace at which this occurs can vary significantly. Given how little was known about the new coronavirus in late 2019 and early 2020, scientists had no choice but to sit and wait.
Fortunately, by the spring, it had become apparent that SARS-CoV-2’s genome was relatively stable. Like the SARS and MERS coronaviruses, the novel virus was confirmed to encode a proof-reading polymerase that limits errors during genome replication (2). The news was a relief to the scientific and public health communities. However, the threat of mutation was not a closed case. As the outbreak continued to grow exponentially through the spring of 2020, SARS-CoV-2 was given an ever-increasing opportunity to mutate. By the summer, multiple point mutations were repeatedly cropping up in genomic analyses – of which a large proportion occurred in the Spike gene (3). The significance of this was unclear at the time, but one mutant quickly gained widespread attention.
First identified in China in January 2020 (4), D614G seeded the early outbreak in Europe and rapidly proliferated – establishing itself as the global consensus (5). After the extent of its proliferation was recognized, it became a contentious point of debate, with multiple groups arguing that its success was either down to chance, or the result of a beneficial phenotype (6). Moreover, given that its Spike protein mediates a range of key functions, including host-cell entry and cell tropisms, changes in its gene sequence were understood to be particularly relevant to infectivity, clinical manifestation, and immune escape.
By late summer, it had become clear that D614G did not pose a significant threat. Though some mutations improved spike flexibility to facilitate stronger binding with the human ACE2 receptor (7), they only conferred a slight increase in affinity for it. What’s more, it only resulted in an even smaller increase in infectivity. Given that the mutation does not occur in Spike’s receptor-binding domain (RBD) – the region responsible for ACE2-binding and the major target of neutralizing antibodies – it fails to mediate resistance to host immunity.
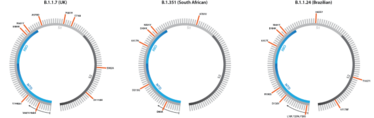
Figure 1. Spike gene sequences for the B.1.1.7, B.1.351, and B.1.1.24 variants, including both point mutations and deletions.
The changing mutational landscape
Since the summer of 2020, the emergence of new mutations has continued to accelerate as global cases have risen at a near-exponential rate (8). This culminated in late 2020 when three new lineages were detected all within the space of a month: B.1.1.7 in the UK (9), B.1.351 (501Y.V2) in South Africa (10), and B.1.1.24 in Brazil (11) (see Figure 1). What’s surprising about the emergence of these variants, in particular, is that each possesses a broad constellation of mutations not seen together before, marking a more significant leap in viral evolution.
Coinciding with their emergence, the countries that detected them witnessed surges in case numbers, potentially associated with increased variant infectivity (12). Given that the promise of vaccine-mediated herd immunity was still many months away, this didn’t come as welcome news, and governments have since had to re-impose strict lockdowns and travel restrictions to contain variant transmission. As vaccination programs progress, transmission is expected to decrease. However, the proportion of vaccine coverage needed to achieve herd immunity is not yet clear, and the bar may rise higher and higher if new variants are more transmissible in naïve and immunized individuals.
To characterize the origin and impact of these mutations, the scientific community has been hard at work. So far, it appears that the increased infectivity of these variants is largely conferred by mutations in the receptor binding domain of their Spike proteins – most notably, E484K and N501Y, as well as the combinatorial N419K:E484K:N501Y, which have all shown to improve affinity for ACE2 (13). All three have emerged repeatedly and independently of one another, suggesting they are advantageous low hanging fruit for the virus to adopt, alongside a string of other, less significant mutations (14). In addition, Spike has shown a tendency to lose portions of its N-terminal domain (NTD) during replication, which, unlike point mutations, cannot be corrected by its proof-reading exonuclease. These deletions have also shown to reoccur and represent a viable means for SARS-CoV-2 to develop more drastic changes (15). The H69/V70 deletion found in the B.1.1.7 (UK) variant, for example, has shown to contribute to increased infectivity, as well as potentially reducing the sensitivity of PCR tests (16).
The second major area of investigation is immunity. As nearly all vaccines that are currently approved or under development target the Spike protein of the original Wuhan-Hu-1 strain, there is concern that mutations could render them less effective. This was first supported by early in vitro studies, showing that the mutations of some variants make them less susceptible to the neutralizing ability of specific antibodies, or are able to escape them entirely (13, 17, 18). Data from South African trials of the Novavax and Johnson & Johnson vaccines also showed slight decreases in vaccine-induced protection, likely due to the locally-circulating variant (19, 20, 21). Fortunately, as the majority of vaccines provoke an immune response to the entire Spike protein, it is expected that the antibody diversity elicited from vaccination will retain at least a moderate degree of protective efficacy, especially against severe COVID-19. The reason for this is because the reduced affinity for one antibody is often compensated for by antibodies that target other regions of the Spike protein, not to mention the oft-forgotten role of cell-mediated immunity. Finally, it’s also worth considering that evolution tends to result in trade-offs, in which the enhancement of one trait, such as receptor binding, is negated by a loss of fitness elsewhere, which is exemplified by D614G being more susceptible to neutralizing antibodies.
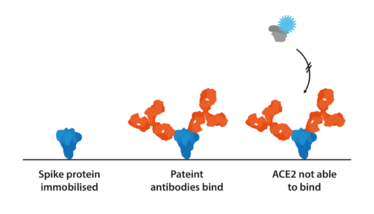
Figure 2. The basic mechanism of a competitive binding assay. If a patient produces neutralizing antibodies, they will preclude the binding of ACE2 with the label, resulting in a negative signal.
Keeping Tabs
The emergence of new SARS-CoV-2 mutations in recent months has driven home the notion that viral evolution – which has otherwise had little impact on the COVID-19 pandemic – could yet result in some last-minute surprises. Mutations are a wake-up call for public health and require systemic surveillance, tracking, and post-vaccination studies. Next-generation sequencing is currently playing a crucial role in surveillance, helping to identify new mutations that occur and track their geographical spread. Supported by epidemiological modeling, NGS provides early indications of whether mutants are fixing in a population, and if they might be more infectious, virulent, or resistant to vaccines.
Prospective studies have also been using techniques such as deep mutational scanning, in which every single possible point mutation is evaluated, as well as predictive computational simulations of binding interactions (21, 22, 23, 24). However, as mutations are additive in their effects, experimental data is essential to inform any changes in the correlates of protection. Furthermore, the leading hypothesis for the origin of recent variants is that immunocompromised individuals incubated unusually long infections, producing low titers of narrow antibody repertoires, that allowed SARS-CoV-2 to mutate (25). It is, therefore, crucial to quantitatively and qualitatively assess the antibody profiles of at-risk groups who have developed natural and vaccine-induced immunity to determine the factors associated with patient-to-patient variation (16).
Neutralization assays have been key in assessing the protective efficacy of patient antibodies and can also be used in conjunction with serial passage to investigate selective pressures. To determine specific affinities and how mutations contribute to protein interactions, assays can be designed to investigate the binding of Spike, ACE2, and monoclonal or polyclonal antibodies. Competitive assays, for example, allow the simultaneous evaluation of seroreactivity against the binding affinity of Spike variants (26) (see Figure 2).
In the case that the protective efficacy of vaccines against moderate-to-severe disease is lowered significantly by new variants, there will be a need to reformulate existing vaccines. Longer-term, there has also been speculation around whether SARS-CoV-2 will become like the endemic coronaviruses that cause seasonal infections, similar to the flu. This may require the development of more broadly protective vaccine candidates de novo. However, in either instance, new multiplexed assays will be required that can quantitatively assess cross-protective efficacy. The scientific community is in a better position than ever before to address these problems if and when they arise.
- 1. Zhu et al., “A Novel Coronavirus from Patients with Pneumonia in China, 2019” New England Journal of Medicine. 20;382(8): 727 (2020).
- 2. Robson et al., “Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting,” Molecular Cell. 79(5):710 (2020).
- 3. Zhan et al., “Molecular Evolution of SARS-CoV-2 Strucutural Genes: Evidence of Positive Selection in Spike Glycoprotein,” BioRxiv (2020).
- 4. Isabel et al., “Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide,” Scientific Reports,10, 14031 (2020).
- 5. Yurkovetskiy et al., “Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant,” Cell (2020).
- 6. Korber et al., “Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus,” Cell, 182, 812 (2020).
- 7. Yurkovetskiy et al., “SARS-CoV-2 Spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain,” BioRxiv (2020).
- 8. World Health Organization Coronavirus (COVID-19) Dasboard. Accessed 2021: https://covid19.who.int/
- 9. NR Faria et al., “Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings” (2021). Available at https://bit.ly/3fKr4Sh.
- 10. A Rambaut et al., “Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations” (2020). Available at https://bit.ly/3wyCZIT.
- 11. H Tegally et al., “Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa,” MedRxiv (2020). https://doi.org/10.1101/2020.12.21.20248640
- 12. JF Cadena et al., “Detection of the new SARS-CoV-2 variant B.1.526 with the Spike E484K mutation in South America” (2021). Available at https://bit.ly/3moeVUH
- 13. Z Wang et al., “mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants,” Nature (2021). https://doi.org/10.1038/s41586-021-03324-6
- 14. P Priya and A Shanker, “Coevolutionary forces shaping the fitness of SARS-CoV-2 spike glycoprotein against human receptor ACE,” Infection, Genetics and Evolution, 87 (2021).
- 15. KR McCarthy et al., “Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape,” Science, 371, 1139 (2021).
- 16. FDA, “Genetic Variants of SARS-CoV-2 May Lead to False Negative Results with Molecular Tests for Detection of SARS-CoV-2 - Letter to Clinical Laboratory Staff and Health Care Providers” (2021). Available at https://bit.ly/3eyPE7Q.
- 17. Z Liu et al., “Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization,” BioRxiv (2020).
- 18. WF Garcia-Beltran et al., “Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity,” MedRxiv (2021).
- 19. Biospace, “Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial” (2021). Available at https://bit.ly/3mlSyPs.
- 20. Johnson & Johnson, “Johnson & Johnson Announces Single-Shot Janssen COVID-19 Vaccine Candidate Met Primary Endpoints in Interim Analysis of its Phase 3 ENSEMBLE Trial” (2021). Available at https://bit.ly/31QLEZi.
- 21. TN Starr et al., “Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding,” Cell, 182,1295 (2020).
- 22. AJ Greaney et al., “Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition,” Cell Host & Microbe, 29,44 (2021).
- 23. MH Cheng et al., “Impact of South African 501.V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-based Computational Assessment,” BioRxiv (2021).
- 24. Sztain et al., “A glycan gate controls opening of the SARS-CoV-2 spike protein,” BioRxiv (2021).
- 25. SM Kissler et al., “Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2,” MedRxiv (2021).
- 26. JR Byrnes et al., “Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding,” Clinical Science and Epidemiology, 5 (2020).
Andy studied pathobiology at the University of Reading and completed a PhD in antibody immunotherapy at the University of Southampton. He then joined the NHS and led a monoclonal antibody research group working in leukemia and lymphoma diagnostics. Andy subsequently joined a major monoclonal antibody supplier to lead their new product development. More recently, he was executive director at bioconjugation specialists Innova Biosciences before joining The Native Antigen Company in 2016.